Change Point Regression
This implementation of change point regression was developed by Julian Stander (University of Plymouth) in Eichenseer et al. (2019).
Assume we want to investigate the relationship between two variables,
Let’s generate
set.seed(10) # change the seed for a different sequence of random numbers
n <- 60 # number of total data points
n_shift <- 35 # the data point at which we introduce a change
x <- rnorm(n,0,1) # generate x
y <- rnorm(n,0,0.5) + 0.5 * x # generate y without a change
y[n_shift:n] <- rnorm(length(n_shift:n),0,1) + 1 * x[n_shift:n] + 0.75 # introduce change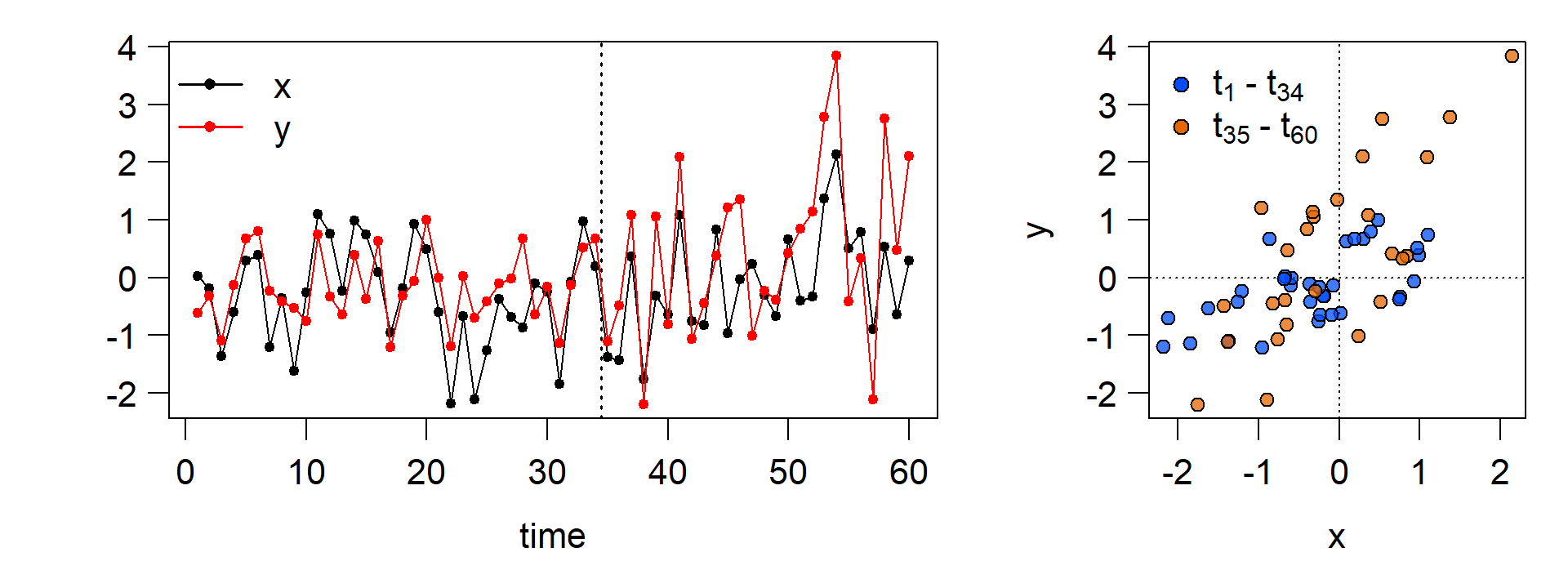
The regression model
Now we build a model that can recover the change point and the linear relationship between
The first part of this model looks like an ordinary least squares regression of
Here we have a single intercept (
From the change point
Implementation in JAGS
Here, we turn to the JAGS programming environment. Understanding a model written for JAGS is not easy at first. If you are keen on learning Bayesian modeling from scratch I can highly recommend Richard McElreath’s book Statistical Rethinking. We will access JAGS with the R2jags package, so we can keep using R even if we are writing a model for JAGS.
Bayesian methods for detecting change points are also available in Stan, as discussed here. An application using English league football data can be found here.
Below, we look at the model. The R code that will be passed to JAGS later is on the left. On the right is an explanation for each line of the model.
We save the model as a function named
model_CPR
Loop over all the data points
note that JAGS uses the precision
of
step takes the value
and
from
back-transform
again, the step function is used to define
We have to define priors for all parameters that are not specified by data.
because
Discrete prior on the change point.
model_CPR <- function(){
### Likelihood or data model part
for(i in 1:n){
y[i] ~ dnorm(mu[i], tau[i])
mu[i] <- alpha_1 +
alpha_2 * step(i - n_change) +
(beta_1 + beta_2 * step(i - n_change))*x[i]
tau[i] <- exp(log_tau[i])
log_tau[i] <- log_tau_1 + log_tau_2 *
step(i - n_change)
}
### Priors
alpha_1 ~ dnorm(0, 1.0E-4)
alpha_2 ~ dnorm(0, 1.0E-4)
beta_1 ~ dnorm(0, 1.0E-4)
beta_2 ~ dnorm(0, 1.0E-4)
log_tau_1 ~ dnorm(0, 1.0E-4)
log_tau_2 ~ dnorm(0, 1.0E-4)
K ~ dcat(p)
n_change <- possible_change_points[K]
}Note that we put priors on
Prepare the data which we pass to JAGS along with the model:
# minimum number of the data points before and after the change
min_segment_length <- 5
# assign indices to the potential change points we allow
possible_change_points <- (1:n)[(min_segment_length+1):(n+1-min_segment_length)]
# number of possible change points
M <- length(possible_change_points)
# probabilities for the discrete uniform prior on the possible change points,
# i.e. all possible change points have the same prior probability
p <- rep(1 / M, length = M)
# save the data to a list for jags
data_CPR <- list("x", "y", "n", "possible_change_points", "p") Load the R2jags package to access JAGS in R:
library(R2jags) Now we execute the change point regression. We instruct JAGS to run three seperate chains so we can verify that the results are consistent. We allow 2000 iterations of the Markov chain Monte Carlo algorithm for each chain, the first 1000 of which will automatically be discarded as burn-in.
CPR <- jags(data = data_CPR,
parameters.to.save = c("alpha_1", "alpha_2",
"beta_1","beta_2",
"log_tau_1","log_tau_2",
"n_change"),
n.iter = 2000,
n.chains = 3,
model.file = model_CPR)The results
To visualise the results and inspect the posterior, we are using the ggmcmc package, which relies on the ggplot2 package. For brevity, we just look at the
library(ggmcmc)
CPR.ggs <- ggs(as.mcmc(CPR)) # convert to ggs object
ggs_traceplot(CPR.ggs, family = "n_change") 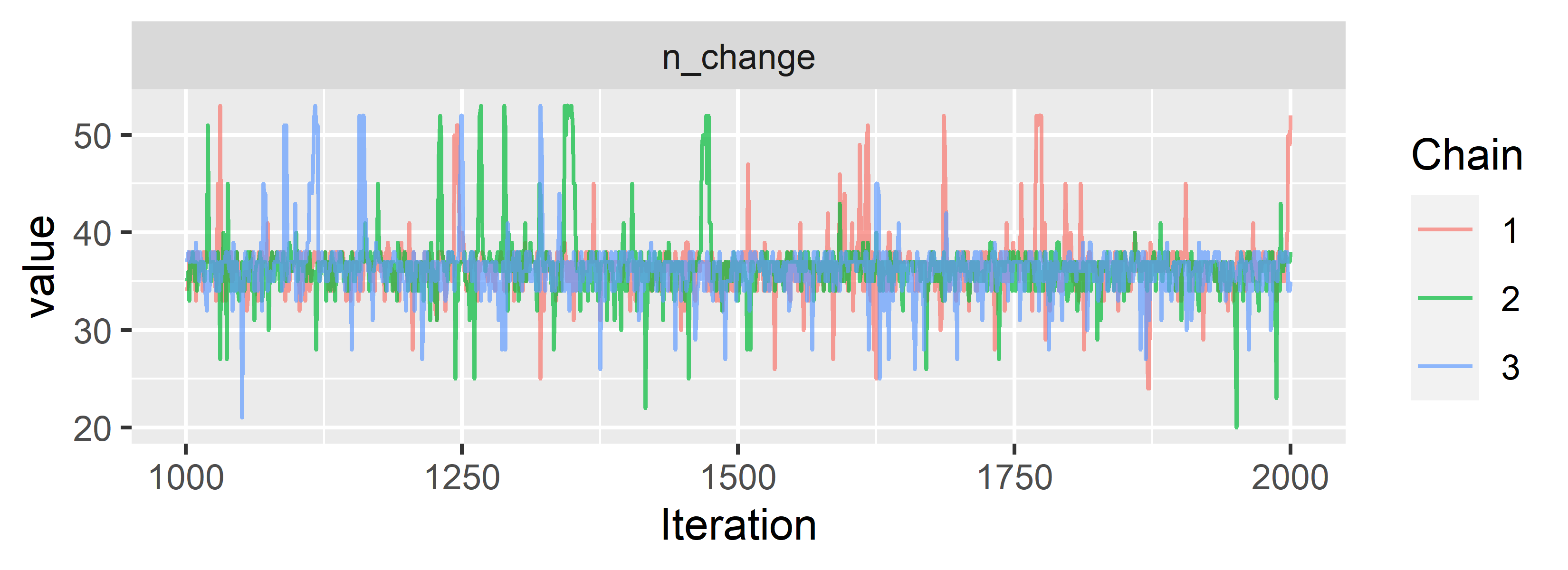
Looks like the chains converge and mix nicely. We can already see that our model locates the change point somewhere between
Let’s look at the posterior probabilities for the possible change points:
ggplot(data = CPR.ggs %>% filter(Parameter == "n_change"),
aes(x=value, y = 3*(..count..)/sum(..count..), fill = as.factor(Chain))) +
geom_vline(xintercept = 35,lty = 2) + geom_bar(position = "identity", alpha = 0.5) +
ylab("posterior probability") + xlab("n_change") + labs(fill='Chain')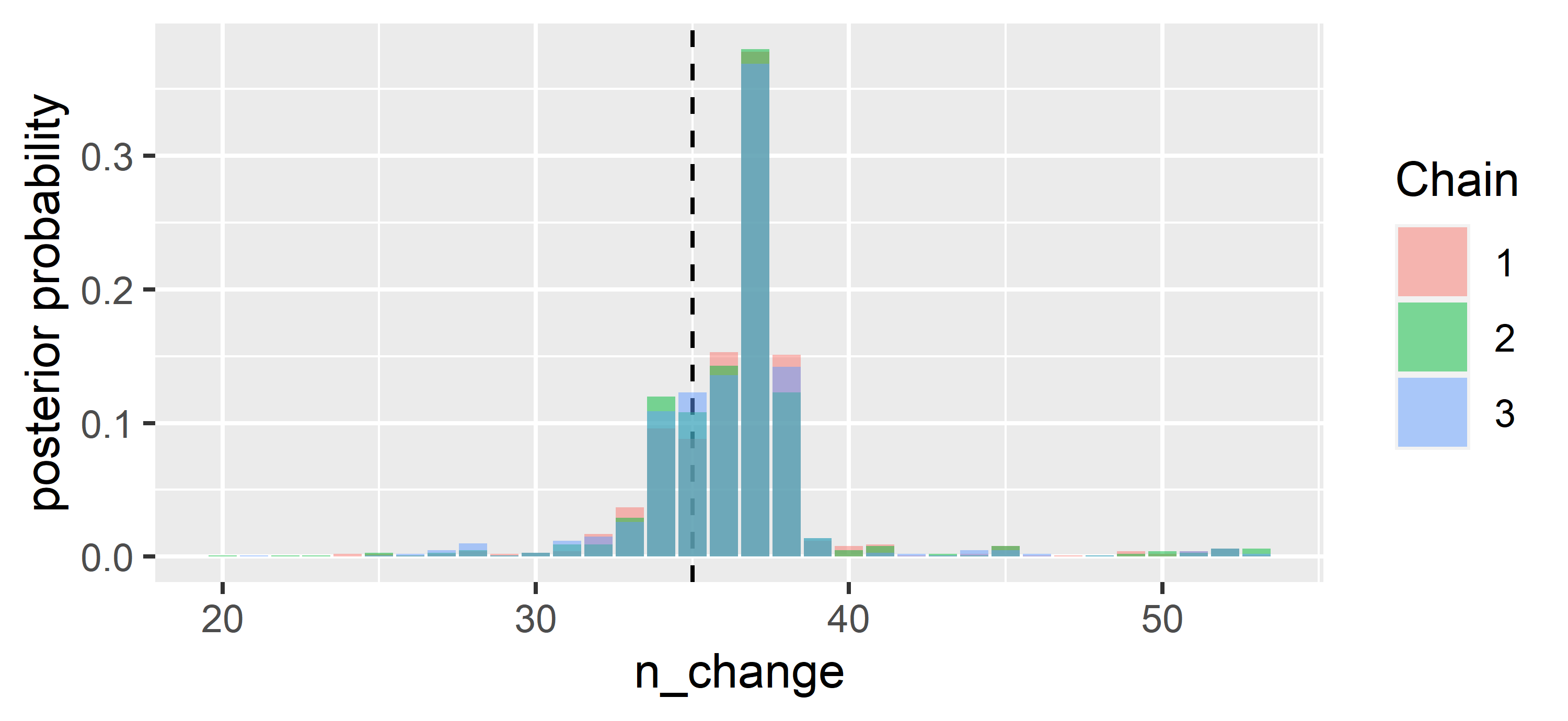
The
Using the posterior distribution, we can answer questions like: “In which interval does the change point fall with 90 % probability?”
quantile(CPR$BUGSoutput$sims.list$n_change, probs = c(0.05, 0.95))## 5% 95%
## 33 39We can also inquire about the probability that the change point falls in the interval
round(length(which(CPR$BUGSoutput$sims.list$n_change %in% 34:38))/
(CPR$BUGSoutput$n.sims),2)## [1] 0.87Finally, let’s have a look at the regression parameters and plot the resulting regressions before and after the most likely change point.
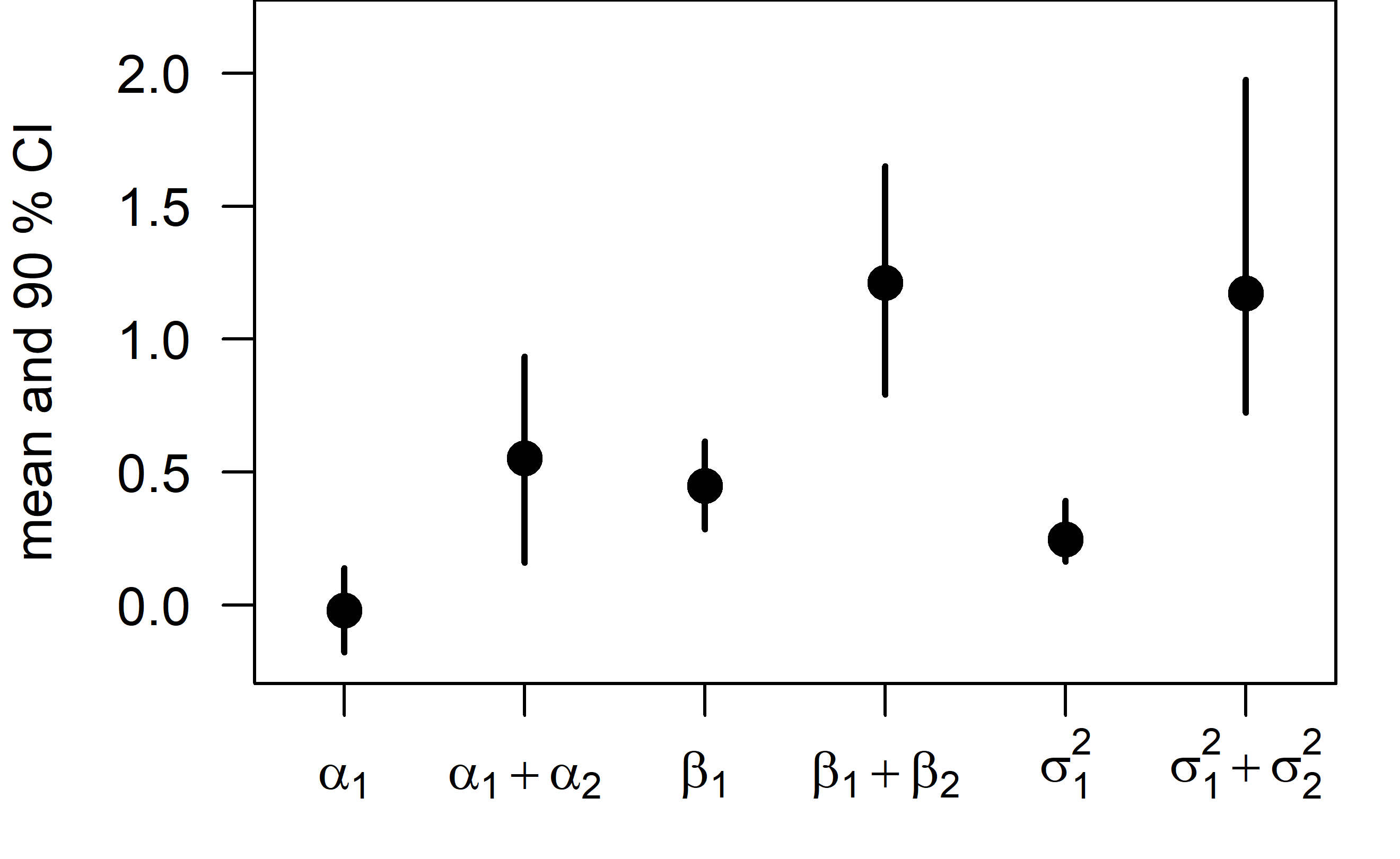 The intercept, slope, and residual variance all increase after the change point.
The intercept, slope, and residual variance all increase after the change point.
This can be immediately seen when plotting the change point regression:
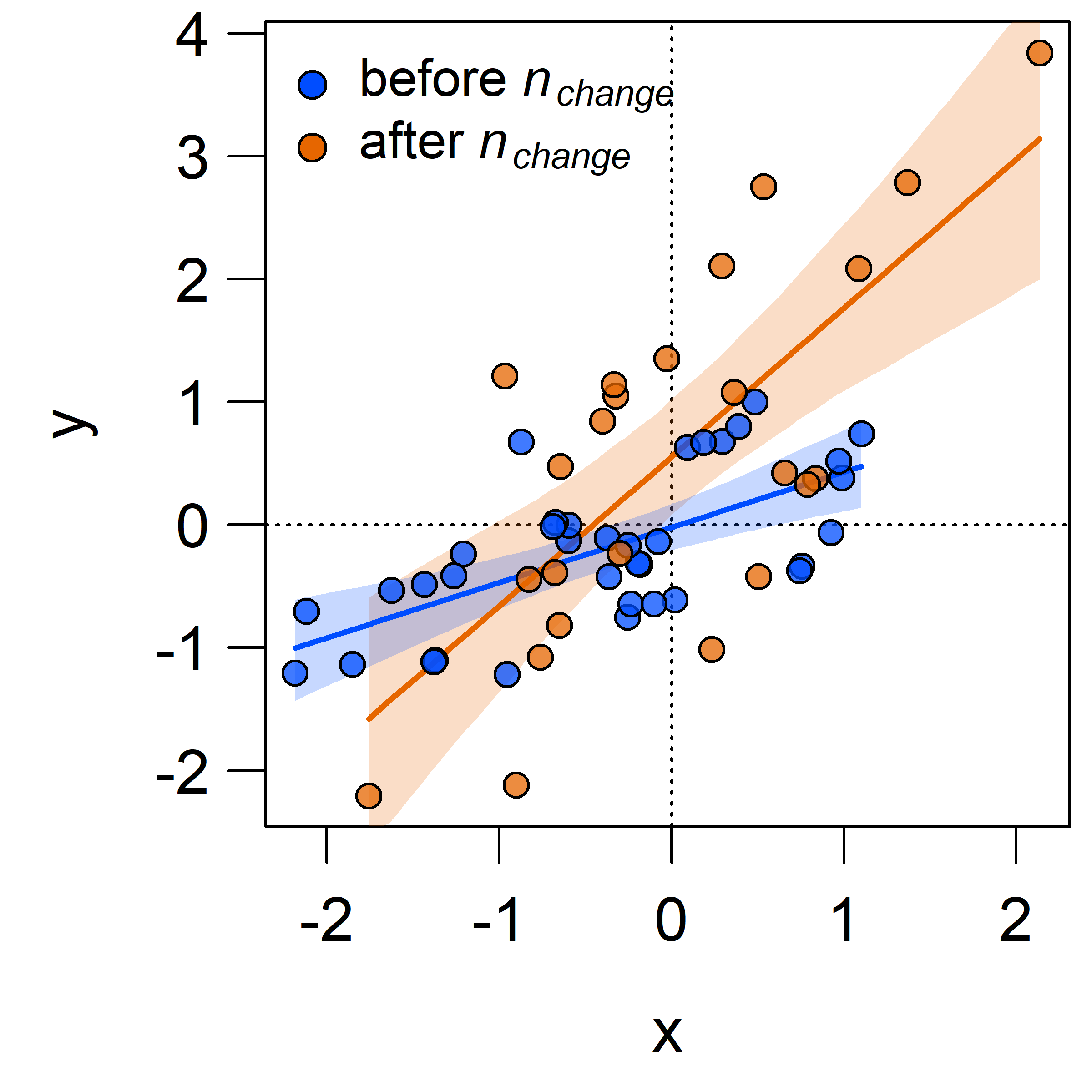 The shaded areas denote
The shaded areas denote
You can find the full R code for this analysis at https://github.com/KEichenseer/Bayesian-Models/blob/main/01-Change_point_regression_with_JAGS.R
Get in touch if you have any comments or questions!